Protein Interactions and Gluten
Abstract
This paper has discussed the distinct types of protein interactions. These were noted to be inclusive of generally the transient interactions and stable interactions. The paper has noted that either of the interactions can be strong or weak. The paper has detailed that the binding parts can either be small or large binding surfaces. Also, they can either be made up of a few peptides long or span of numerous amino acids. The paper has dug into the biological effects of the protein-protein interactions that involve various functional objectives. For those seeking healthcare dissertation help, understanding these interactions can provide insights into various functional outcomes. The paper has also discussed the gluten as a component of wheat amongst other cereals. Gluten has been noted to be a family protein that is found in proteins. The two proteins that are inclusive of the glutenin and the gliadin have been noted to be part of gluten. The paper has described and discussed the proteins. They have been stated to be responsible for the adverse health effects of gluten. The proteins have been noted to diversely play an important role in determining the quality of baking of wheat. The paper has explored the mechanisms of gluten development during the processing and the characteristics that gluten confers to the dough and finished product.
Introduction
Proteins are macromolecules that are formed by many amino acids coming together to form bonds. They are large and complex molecules which are essential to the body in diverse ways. Most of the works in the cells are done by the proteins. Besides, proteins form the structure of the cells. They are crucial to the function and regulation of the tissues and organs of the body through enzymes that carry out almost all of the chemical reactions that occur in cells. Through protein-protein interactions, the molecules are of inevitable significance to man. Consequently, this essay aims to discuss the mechanisms that underlie the interactions and then exploring the individual proteins that are present in gluten and thus important to the baking quality of wheat.

The Mechanisms behind Protein-Protein Interactions
Proteins can be termed as the macromolecules that are comprised of amino acids. They are large in size and are polymers of the structural subunits known as the amino acids. 20 distinct amino acids are in existence in proteins while several of these amino acids are interlinked to each other in long chains to form the proteins. There are various types of protein-protein interactions. According to Brito & Pinney (2017), proteins exhaustively control biological systems in a cell. While many proteins are independent, a good number interact with others for efficient biological activity. In understanding the protein function and the biology of the cell, it is of utmost significance to categorize proteins based on strategies that are inclusive of co-immunoprecipitation, pull-down-assays, crosslinking, label transfer, and far-western blot analysis is crucial, notes Brito & Pinney (2017). The interactions alter the properties of enzymes that lead to subtle changes in the substrate allosteric effects. Besides, the interactions allow for substrate channeling and create new binding sites. Also, inactivation of proteins has been noted as a result of the protein-protein interactions. Lastly, the interactions serve a regulatory role. Leucine zipper, as a surface domain, offers the capability of protein interactions through alpha helices that link to each other through the hydrophobic bonding. Due to the tight molecular packing, the domain provides a stable binding for multi-protein complexes. The hydrogen bonding, salt bridges and the van der Waals forces all contribute to the linking of proteins to each other. Proteins enable most biological processes such as gene interactions, cell growth, proliferation, nutrient uptake, morphology, motility, intercellular communication, and apoptosis within a cell. Protein expression has been termed as a dynamic process by various scholars and most of them are presented in a cell-type dependent manner.
Protein associations are notable as stable and transient and can be strong or weak. Stability in protein interactions are is due to multi-sub unit complexes of pure proteins which can be the same or different (Sinz, 2018). For example, stable complexes are drawn from hemoglobin and the core RNA polymerase multi-sub unit interactions. On the other hand, transient interactions control the majority of the cellular process of the body (Brito & Pinney, 2017). They are temporary in nature and need standard conditions that promote interaction like addition of phosphorus, changes in conformation, and localization discrete areas of the cells which can all be strong or weak, fast or slow (Sinz, 2018). Transiently interacting proteins are involved in various cellular processes that are including the protein modification process, transport, folding, signaling, apoptotic and anti-apoptotic processes.
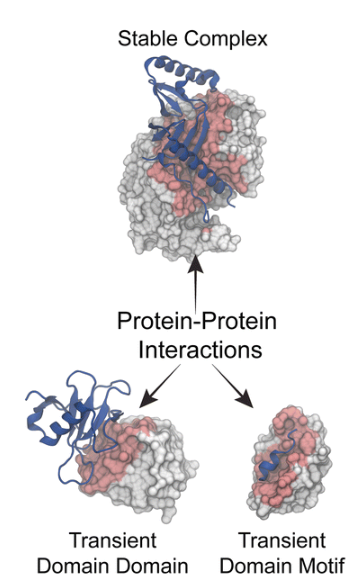
Protein-protein interactions significantly contribute to the efficiency of various cell processes. (Brito & Pinney, 2017). The formation of the protein complex is equally contributes to formulation of oligomer enzyme’s active sites. Besides, they have a contribution in the maintenance of their effective conformation. Besides, it is crucial to the various regulatory processes that include the signal transduction, electron transport system, DNA synthesis, intracellular structures formation, and the antigen-antibody reactions (Sinz, 2018).
There are various mechanisms that underlie the protein-protein interaction. These mechanisms are basically based on the various forces that are involved in the interaction steric, hydrophobic, electrostatic interactions and hydrogen bonds (Brito & Pinney, 2017).
Hydrophobic Interaction
The hydrophobic force contributes significantly to protein-protein interactions. Various studies have demonstrated this interaction in a broad context. According to Researchers, permanent complexes have higher contribution of hydrophobic interactions relative to the non-obligate protein complexes (Maniaci & Ciulli, 2019). For Permanent complexes, they are mostly in a bound state which a high contribution of hydrogen bonds. Non-obligate complexes are energetically unfavorable due to the hydrophobicity of their surfaces that results from the fact that they are assembled in the water environment for a relatively shorter time. Other non-obligate complex of membrane proteins are formed by hydrophobic interactions (Maniaci & Ciulli, 2019). In an interaction of enzymes with peptide inhibitors substrates, the contacted interfaces can have the hydrophilic surfaces (Brito & Pinney, 2017). In the contact surfaces, the hydrophobic regions are organized as patches. These patches vary in number from 1 to 15. The sizes of the patches are within 200-400 A2 (Schleinitz et al., 2019). The patches have been noted to overlap lowly.
Electrostatics
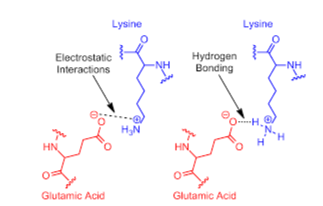
This is another force that is involved in protein-protein interactions. On the interacting surfaces, charges are located complementarily to each other (Maniaci & Ciulli, 2019). Lately, studies note that the electrostatic complementarity of the interacting proteins surfaces. Studies have further noted that the charge density varies (0-12) for groups that are charged in each interface surface (Brito & Pinney, 2017). Salt bridges have been noted across the distribution of the charges in the interfaces of the area that is interacting.. These are favorable interactions with other charges as well as the hydrophilic residues within their surroundings. This also enhances the complex formation rate. The formation of protein interactions are promoted by the long range of electrostatic forces (Schleinitz et al., 2019). Electrostatic interactions define the lifetime of protein complexes.
Hydrogen Bonding
The area of the sub unit surfaces are determined by the average number of the hydrogen bonds. In protein-protein interactions, hydrogen bonds are mostly found to be of the of oxygen and nitrogen types. Most hydrogen bonds are constituted of side chains of amino acids as have been proven by experiments on the structure of proteins (Schleinitz et al., 2019). However, the beta sheet interfaces are exempted. Besides, research has indicated that protein complexes with peptide inhibitors or substrate have a chain that basically form hydrogen bonds which are never in an optimal position and are usually weak due to this fact (Schleinitz et al., 2019). The interactions between the hydrogen bonds of protein and water are good interactions unlike the hydrogen bonds between protein surfaces themselves. Water molecules interact with protein group to form a connection in the protein domains.
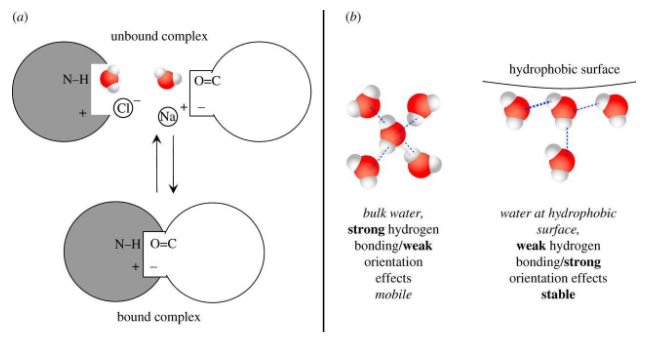
Water Interfaces
At the complex interfaces, water molecules are very popular. The number of water molecules at the complexes interfaces varies from 1 to 50. The molecules surround the contacting surfaces (Schleinitz et al., 2019). Either way, they are buried in the contacting surfaces within the protein surface cavities in a highly coordinated manner (Maniaci & Ciulli, 2019). Hydrogen has been observed between the connection of water molecules and protein groups forming aqueous networks in the protein domains. Notably, the interface water molecules have been found to stabilize the various protein complexes that they are involved in through the formation of extra hydrogen bonds. They also interact with charges to increase the shape in addition to charge complementarity (Brito & Pinney, 2017). The protein-protein binding is associated with partial desolvation of the contacted surfaces that is more in complexes that has a weakly charged or neutral reactant.
Steric Complementarity
The interface surfaces of proteins complete each other depending on the particular type of the protein interaction. Permanent complexes exhibit highest complementarity while non-obligate complexes and Protein inhibitors show a lower complementarity and antigen-antibody complexes exhibiting the worst complementarity (Maniaci & Ciulli, 2019). The Protein interfaces contain cavities that represent a tenth of interfaces mostly filled with solvents (Maniaci & Ciulli, 2019).
Conformation
The distinct structural changes that occur during the formation of complexes have been detailed by various researchers. The changes cause induced-fit effects (Schleinitz et al., 2019). Protein-protein interactions affect the structure of the individual proteins in diverse ways that include changes in the positions of the side chain amino acids, motion of the main chain or even the domain (Errington et al., 2019). Some amino acids have been noted to deviate due to the conformational changes as a result of the binding initiated by lysozyme of the distinct antibodies. Low-energy conformational changes allowing hydrogen bond formation and the packing of the amino acid residues result in the rearrangement in the protein backbone (Schleinitz et al., 2019).
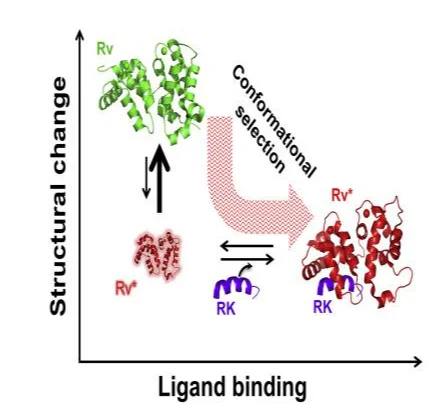
During the assembling of the non-obligate complexes, protein folding occurs alongside optimization of the subunit skeletons impacting on the formation of the cavities in the presence of water molecules at the complex interfaces, non-optimal hydrogen bond geometry (Errington et al., 2019).
This Proteins Involved In Gluten, the Mechanisms of Gluten Development during Processing and the Characteristics That Gluten Confers To Dough and Finished Product
Research has noted that gluten significantly impact on the quality of baking noted in wheat (Marti & Pagani, 2013). Basically, gluten is a combination of various proteins that can be categorized bialy on the basis of solubility in aqueous alcohols, notably, the soluble gliadins and the insoluble glutenins (Biesiekierski, 2017).
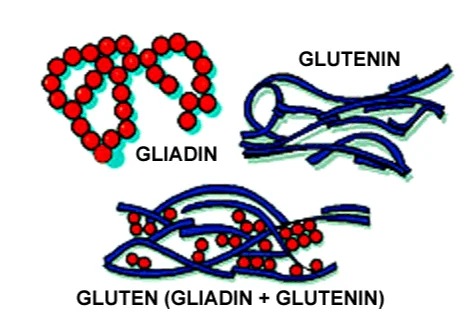
The two categories each constitute of closely associated protein constituents high in glutamine and proline contents. Gliadins are monomeric proteins that are classified as alpha, beta, gamma, or omega proteins (Biesiekierski, 2017). The glutenin fractions aggregated proteins with different sizes and connected via interchain disulphide bonds. Basically, the glutenin and the gliadins are the main proteins that are present in gluten (Marti & Pagani, 2013). Gliadin, just like glutenin, is a component of gluten important in giving bread the ability to rise effectively during baking (Biesiekierski, 2017).
Glutenin is a protein aggregate of relatively higher molecular mass and lower molecular mass subunits. The protein is not only responsible for the strength but also the elasticity of dough (Wieser, 2017). The two proteins differ in the aspect that gliadin is water insoluble while glutenin is water soluble.
Gluten is obtained from the mixing of flour with water and starch washed out (Biesiekierski, 2017). The gliadin forms the compact spherical shape when it folds while the glutenin assumes a more elongated rope shape when folded. The two proteins have hydrogen bonds between them due to the amino acids that they possess (Marti & Pagani, 2013). The two proteins form the network within raised dough that enables the dough in maintaining the shape during baking (Wieser, 2017). When the bread dough is kneaded, the network is formed. This results in the stretching out of the glutenin proteins and aligning them with one another (Naqash et al., 2017). The fresh bonds between the gliadin and the glutenin molecules are responsible for preserving the new shape besides reinforcing the entire protein network (Marti & Pagani, 2013). The network allows the bread to emerge from the oven as a light and fluffy loaf (Biesiekierski, 2017). The network is made up of aligned and elongated strands that enable the expansion of the internal structure of bread.

Conclusion
The paper has discussed the protein-protein interactions as well as their types and properties. The binding parts of the proteins have also been explored and discussed in a broad context. Also, the paper has explored the biological effects of the protein-protein interactions that involve various functional objectives. The paper has also discussed the gluten as a component of wheat amongst other cereals. Gluten has two proteins that are inclusive of the glutenin and the gliadins have been noted to be part of gluten. The paper has described and discussed the proteins. The proteins have been noted to diversely play an important role in determining the quality of baking of wheat. The paper has explored the mechanisms of gluten development during the processing and the characteristics that gluten confers to the dough and finished product. Further research is necessary to understand the interactions better.
Take a deeper dive into Protecting Dementia Patients from Discrimination with our additional resources.
References
Biesiekierski, J. R. (2017). What is gluten?. Journal of gastroenterology and hepatology, 32, 78-81.
Brito, A. F., & Pinney, J. W. (2017). Protein–protein interactions in virus–host systems. Frontiers in microbiology, 8, 1557.
Errington, W. J., Bruncsics, B., & Sarkar, C. A. (2019). Mechanisms of noncanonical binding dynamics in multivalent protein–protein interactions. Proceedings of the National Academy of Sciences, 116(51), 25659-25667.
Jones, S., & Thornton, J. M. (1996). Principles of protein-protein interactions. Proceedings of the National Academy of Sciences, 93(1), 13-20.
Makepeace, K. A., Mohammed, Y., Rudashevskaya, E. L., Petrotchenko, E. V., Vögtle, F. N., Meisinger, C., ... & Borchers, C. H. (2020). Improving Identification of In-organello Protein-Protein Interactions Using an Affinity-enrichable, Isotopically Coded, and Mass Spectrometry-cleavable Chemical Crosslinker. Molecular & Cellular Proteomics, 19(4), 624-639.
Maniaci, C., & Ciulli, A. (2019). Bifunctional chemical probes inducing protein–protein interactions. Current opinion in chemical biology, 52, 145-156.
Marcotte, E. M., Pellegrini, M., Ng, H. L., Rice, D. W., Yeates, T. O., & Eisenberg, D. (1999). Detecting protein function and protein-protein interactions from genome sequences. Science, 285(5428), 751-753.
Marti, A., & Pagani, M. A. (2013). What can play the role of gluten in gluten free pasta?. Trends in food science & technology, 31(1), 63-71.
McCutcheon, D. C., Lee, G., Carlos, A., Montgomery, J. E., & Moellering, R. E. (2019). Photoproximity Profiling of Protein–Protein Interactions in Cells. Journal of the American Chemical Society, 142(1), 146-153.
Naqash, F., Gani, A., Gani, A., & Masoodi, F. A. (2017). Gluten-free baking: Combating the challenges-A review. Trends in Food Science & Technology, 66, 98-107.
Niland, B., &Cash, B. D. (2018). Health benefits and adverse effects of a gluten-free diet in non–celiac disease patients. Gastroenterology & hepatology, 14(2), 82.
Nooren, I. M., & Thornton, J. M. (2003). Diversity of protein–protein interactions. The EMBO journal, 22(14), 3486-3492.
Phizicky, E. M., & Fields, S. (1995). Protein-protein interactions: methods for detection and analysis. Microbiological reviews, 59(1), 94-123.
Schleinitz, M., Sadowski, G., & Brandenbusch, C. (2019). Protein-protein interactions and water activity coefficients can be used to aid a first excipient choice in protein formulations. International journal of pharmaceutics, 569, 118608.
Sheinerman, F. B., Norel, R., & Honig, B. (2000). Electrostatic aspects of protein–protein interactions. Current opinion in structural biology, 10(2), 153-159.
Sijbesma, E., Visser, E., Plitzko, K., Thiel, P., Milroy, L. G., Kaiser, M., ... & Ottmann, C. (2020). Structure-based evolution of a promiscuous inhibitor to a selective stabilizer of protein–protein interactions. Nature communications, 11(1), 1-9.
Sinz, A. (2018). Cross‐linking/mass spectrometry for studying protein structures and protein–protein interactions: where are we now and where should we go from here?. Angewandte Chemie International Edition, 57(22), 6390-6396.
Spreitzer, E., Usluer, S., & Madl, T. (2020). Probing surface accessibility in the dynamic meshwork of protein-protein interactions. Journal of Molecular Biology.
Wei, L., Xing, P., Zeng, J., Chen, J., Su, R., & Guo, F. (2017). Improved prediction of protein–protein interactions using novel negative samples, features, and an ensemble classifier. Artificial Intelligence in Medicine, 83, 67-74.
Wieser, H. (2017). Chemistry of gluten proteins. Food microbiology, 24(2), 115-119.
Zhong, M., Lee, G. M., Sijbesma, E., Ottmann, C., & Arkin, M. R. (2019). Modulating protein–protein interaction networks in protein homeostasis. Current opinion in chemical biology, 50, 55-65.
- 24/7 Customer Support
- 100% Customer Satisfaction
- No Privacy Violation
- Quick Services
- Subject Experts



